Shape the future of Movebank
Please take our user survey! Your feedback is much appreciated.Tracking data
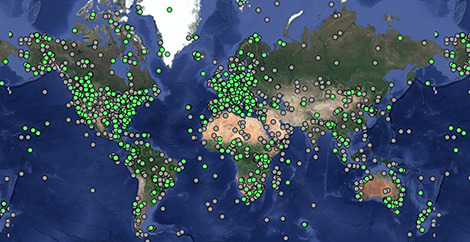
Explore animal movements on our interactive map.
Get started
Are you a researcher, journalist, student or developer? Whether you want to explore what’s here, add your data, or initiate a new project, get headed in the right direction.
News
December 2025 Newsletter
Dear Movebank users,
As 2025 draws to a close, below are some highlights from this past ...
About us
Movebank is a free, online database of animal tracking data hosted by the Max Planck Institute of Animal Behavior. We help animal tracking researchers to manage, share, protect, analyze and archive their data.
- 9.6 billion locations
- 8.3 billion other sensor records
- 9,622 studies
- 1,618 taxa
- 5,046 data owners